Amundsen
PRODMetadata Ingestion
1. Visit the Services Page
Click Settings in the side navigation bar and then Services.
The first step is to ingest the metadata from your sources. To do that, you first need to create a Service connection first.
This Service will be the bridge between OpenMetadata and your source system.
Once a Service is created, it can be used to configure your ingestion workflows.
Select your Service Type and Add a New Service
Add a new Service from the Services page
Select your Service from the list
4. Name and Describe your Service
Provide a name and description for your Service.
Service Name
OpenMetadata uniquely identifies Services by their Service Name. Provide a name that distinguishes your deployment from other Services, including the other Amundsen Services that you might be ingesting metadata from.
Note that when the name is set, it cannot be changed.
Provide a Name and description for your Service
5. Configure the Service Connection
In this step, we will configure the connection settings required for Amundsen.
Please follow the instructions below to properly configure the Service to read from your sources. You will also find helper documentation on the right-hand side panel in the UI.
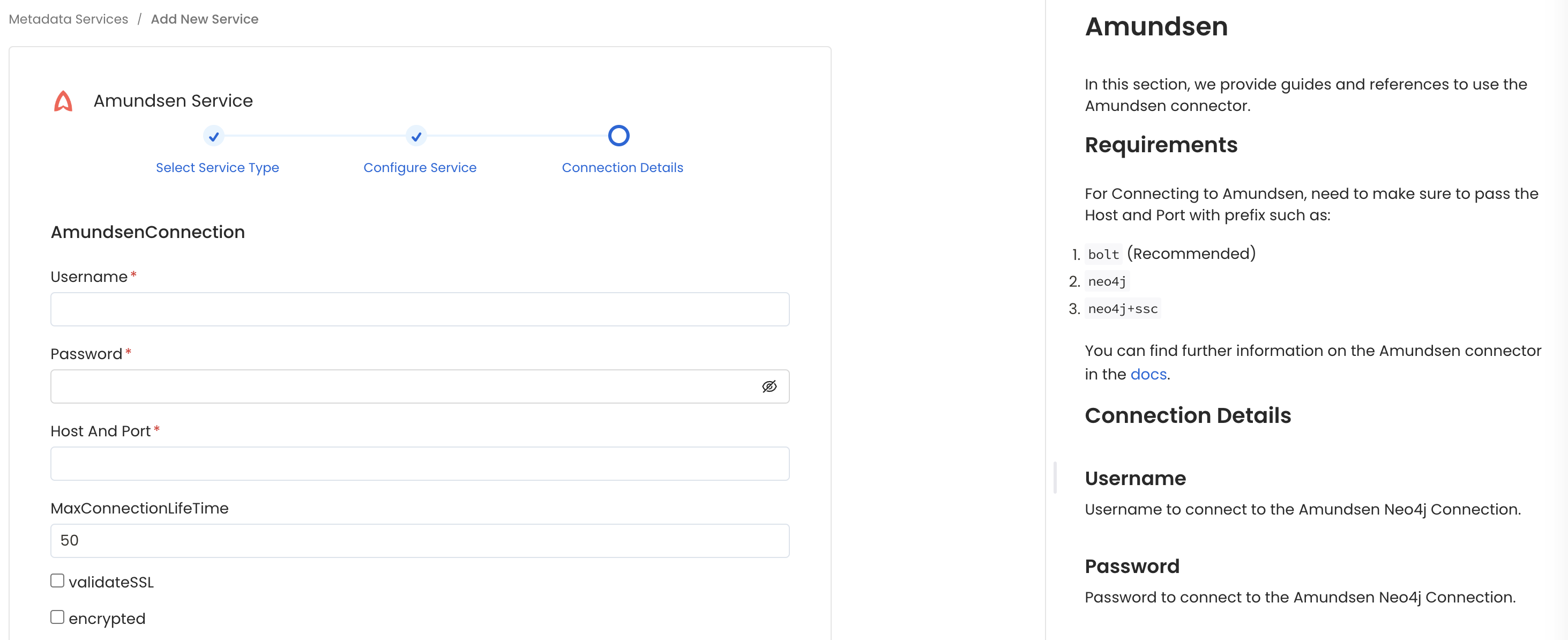
Configure the Service connection by filling the form
Connection Details
When using a Hybrid Ingestion Runner, any sensitive credential fields—such as passwords, API keys, or private keys—must reference secrets using the following format:
This applies only to fields marked as secrets in the connection form (these typically mask input and show a visibility toggle icon).
For a complete guide on managing secrets in hybrid setups, see the Hybrid Ingestion Runner Secret Management Guide.
- username: Enter the username of your Amundsen user in the Username field. The specified user should be authorized to read all databases you want to include in the metadata ingestion workflow.
- password: Enter the password for your amundsen user in the Password field.
- hostPort: Host and port of the Amundsen Neo4j Connection. This expect a URI format like: bolt://localhost:7687.
- maxConnectionLifeTime (optional): Maximum connection lifetime for the Amundsen Neo4j Connection
- validateSSL (optional): Enable SSL validation for the Amundsen Neo4j Connection.
- encrypted (Optional): Enable encryption for the Amundsen Neo4j Connection.
6. Test the Connection
Once the credentials have been added, click on Test Connection and Save the changes.
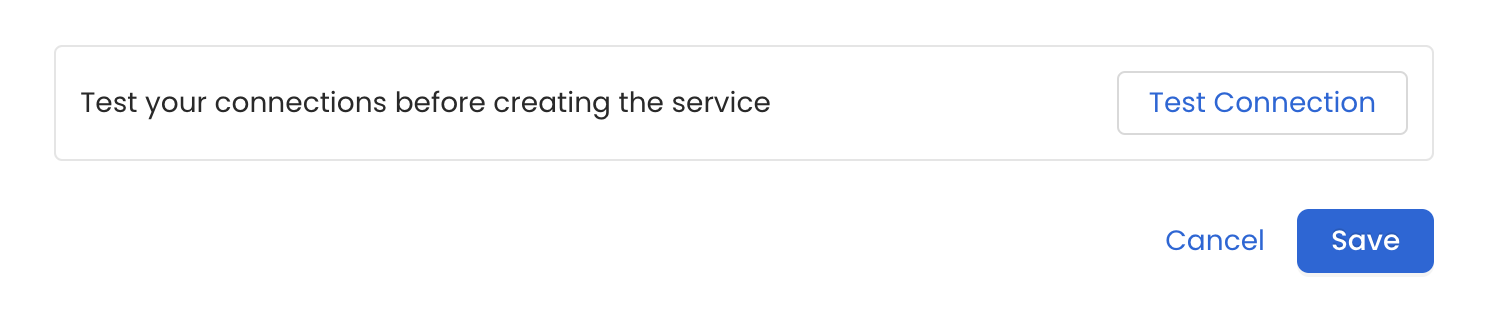
Test the connection and save the Service
7. Configure Metadata Ingestion
In this step we will configure the metadata ingestion pipeline, Please follow the instructions below
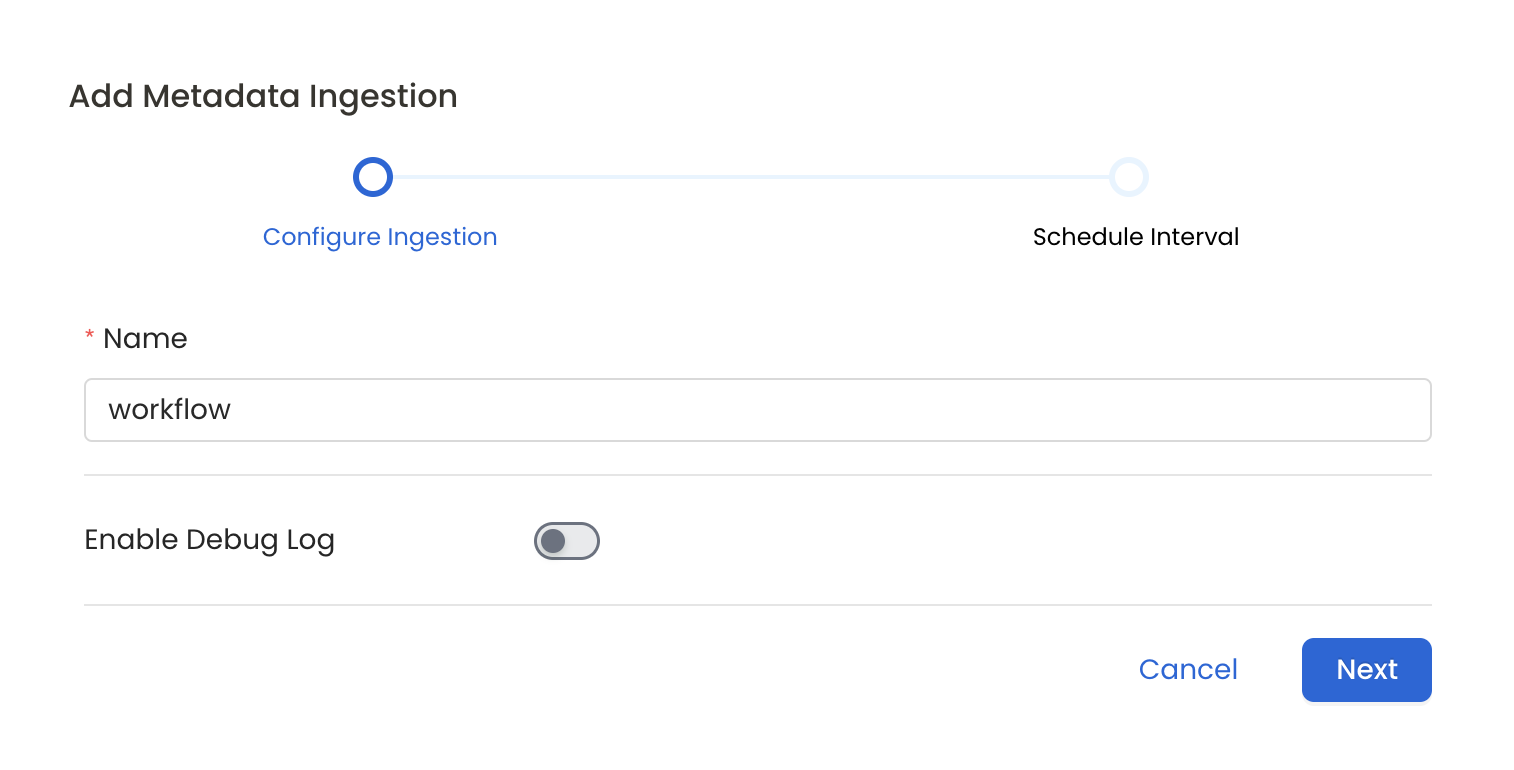
Configure Metadata Ingestion Page
8. Schedule the Ingestion and Deploy
Scheduling can be set up at an hourly, daily, weekly, or manual cadence. The timezone is in UTC. Select a Start Date to schedule for ingestion. It is optional to add an End Date.
Review your configuration settings. If they match what you intended, click Deploy to create the service and schedule metadata ingestion.
If something doesn't look right, click the Back button to return to the appropriate step and change the settings as needed.
After configuring the workflow, you can click on Deploy to create the pipeline.
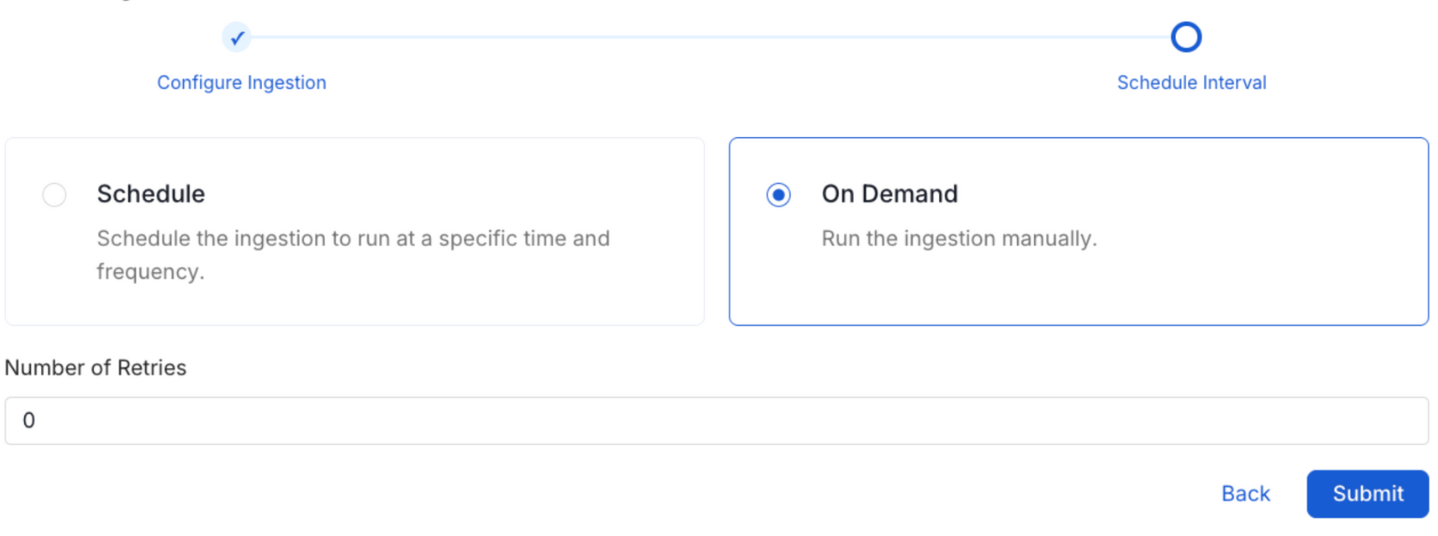
Schedule the Ingestion Pipeline and Deploy
9. View the Ingestion Pipeline
Once the workflow has been successfully deployed, you can view the Ingestion Pipeline running from the Service Page.
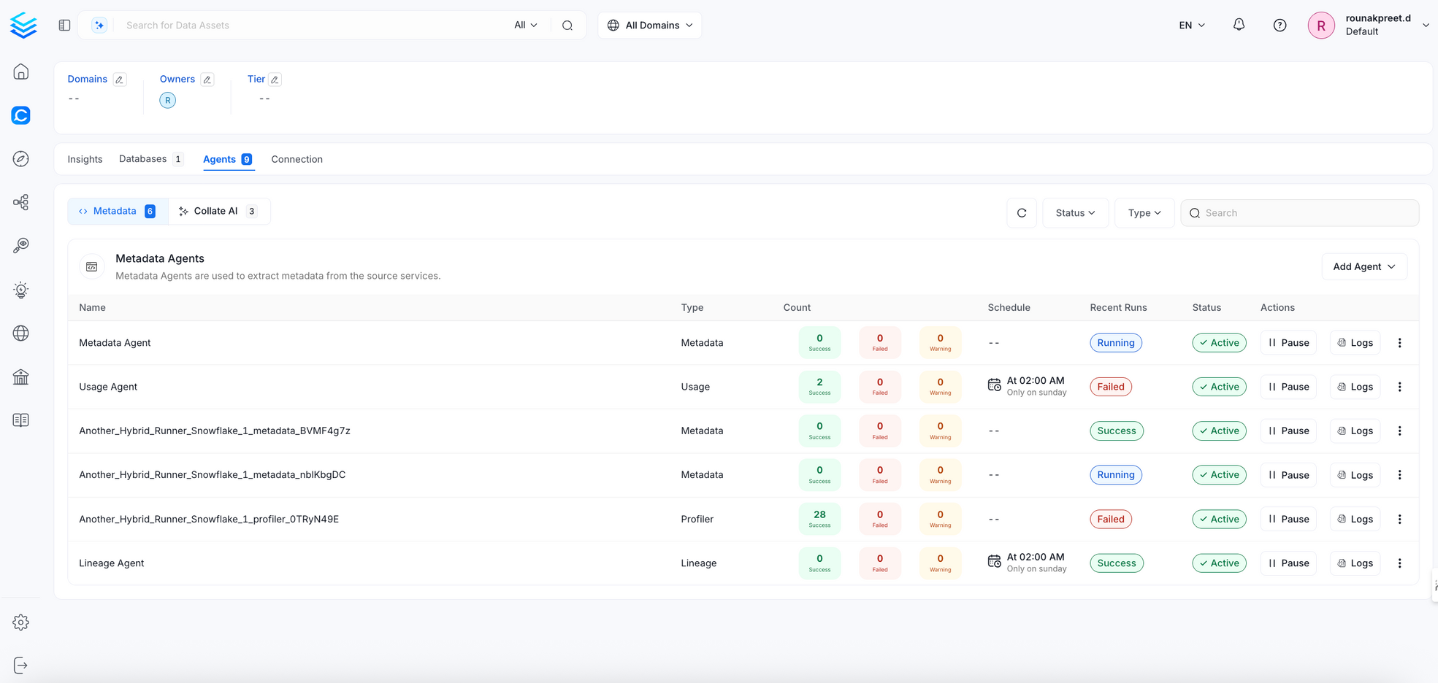
View the Ingestion Pipeline from the Service Page
If AutoPilot is enabled, workflows like usage tracking, data lineage, and similar tasks will be handled automatically. Users don’t need to set up or manage them - AutoPilot takes care of everything in the system.