S3 Storage
PRODThis page contains the setup guide and reference information for the S3 connector.
Configure and schedule S3 metadata workflows from the OpenMetadata UI:
Requirements
We need the following permissions in AWS:
S3 Permissions
For all the buckets that we want to ingest, we need to provide the following:
s3:ListBuckets3:GetObjects3:GetBucketLocations3:ListAllMyBuckets
Note that the Resources should be all the buckets that you'd like to scan. A possible policy could be:
CloudWatch Permissions
Which is used to fetch the total size in bytes for a bucket and the total number of files. It requires:
cloudwatch:GetMetricDatacloudwatch:ListMetrics
The policy would look like:
OpenMetadata Manifest
In any other connector, extracting metadata happens automatically. In this case, we will be able to extract high-level metadata from buckets, but in order to understand their internal structure we need users to provide an openmetadata.json file at the bucket root.
Supported File Formats: [ "csv", "tsv", "avro", "parquet", "json", "json.gz", "json.zip" ]
You can learn more about this here. Keep reading for an example on the shape of the manifest file.
OpenMetadata Manifest
Our manifest file is defined as a JSON Schema, and can look like this:
Entries: We need to add a list of entries. Each inner JSON structure will be ingested as a child container of the top-level one. In this case, we will be ingesting 4 children.
Simple Container: The simplest container we can have would be structured, but without partitions. Note that we still need to bring information about:
- dataPath: Where we can find the data. This should be a path relative to the top-level container.
- structureFormat: What is the format of the data we are going to find. This information will be used to read the data.
- separator: Optionally, for delimiter-separated formats such as CSV, you can specify the separator to use when reading the file. If you don't, we will use
,for CSV and/tfor TSV files.
After ingesting this container, we will bring in the schema of the data in the dataPath.
Partitioned Container: We can ingest partitioned data without bringing in any further details.
By informing the isPartitioned field as true, we'll flag the container as Partitioned. We will be reading the source files schemas', but won't add any other information.
Single-Partition Container: We can bring partition information by specifying the partitionColumns. Their definition is based on the JSON Schema definition for table columns. The minimum required information is the name and dataType.
When passing partitionColumns, these values will be added to the schema, on top of the inferred information from the files.
Multiple-Partition Container: We can add multiple columns as partitions.
Note how in the example we even bring our custom displayName for the column dataTypeDisplay for its type.
Again, this information will be added on top of the inferred schema from the data files.
Automated Container Ingestion: Registering all the data paths one by one can be a time consuming job, to make the automated structure container ingestion you can provide the depth at which all the data is available.
Let us understand this with the example, suppose following is the file hierarchy within my bucket.
all my tables folders which contains the actual data are available at depth 3, hence when you specify the depth: 3 in manifest entry all following path will get registered as container in OpenMetadata with this single entry
saving efforts to add 4 individual entries compared to 1
Unstructured Container: OpenMetadata supports ingesting unstructured files like images, pdf's etc. We support fetching the file names, size and tags associates to such files.
In case you want to ingest a single unstructured file, then just specifying the full path of the unstructured file in datapath would be enough for ingestion.
In case you want to ingest all unstructured files with a specific extension for example pdf & png then you can provide the folder name containing such files in dataPath and list of extensions in the unstructuredFormats field.
In case you want to ingest all unstructured files with irrespective of their file type or extension then you can provide the folder name containing such files in dataPath and ["*"] in the unstructuredFormats field.
Global Manifest
You can also manage a single manifest file to centralize the ingestion process for any container, named openmetadata_storage_manifest.json. For example:
In that case, you will need to add a containerName entry to the structure above. For example:
The fields shown above (dataPath, structureFormat, isPartitioned, etc.) are still valid.
Container Name: Since we are using a single manifest for all your containers, the field containerName will help us identify which container (or Bucket in S3, etc.), contains the presented information.
You can also keep local manifests openmetadata.json in each container, but if possible, we will always try to pick up the global manifest during the ingestion.
Metadata Ingestion
1. Visit the Services Page
The first step is ingesting the metadata from your sources. Under Settings, you will find a Services link an external source system to OpenMetadata. Once a service is created, it can be used to configure metadata, usage, and profiler workflows.
To visit the Services page, select Services from the Settings menu.
Find Dashboard option on left panel of the settings page
Add a new Service from the Storage Services page
Select your service from the list
5. Configure the Service Connection
In this step, we will configure the connection settings required for this connector. Please follow the instructions below to ensure that you've configured the connector to read from your S3 service as desired.
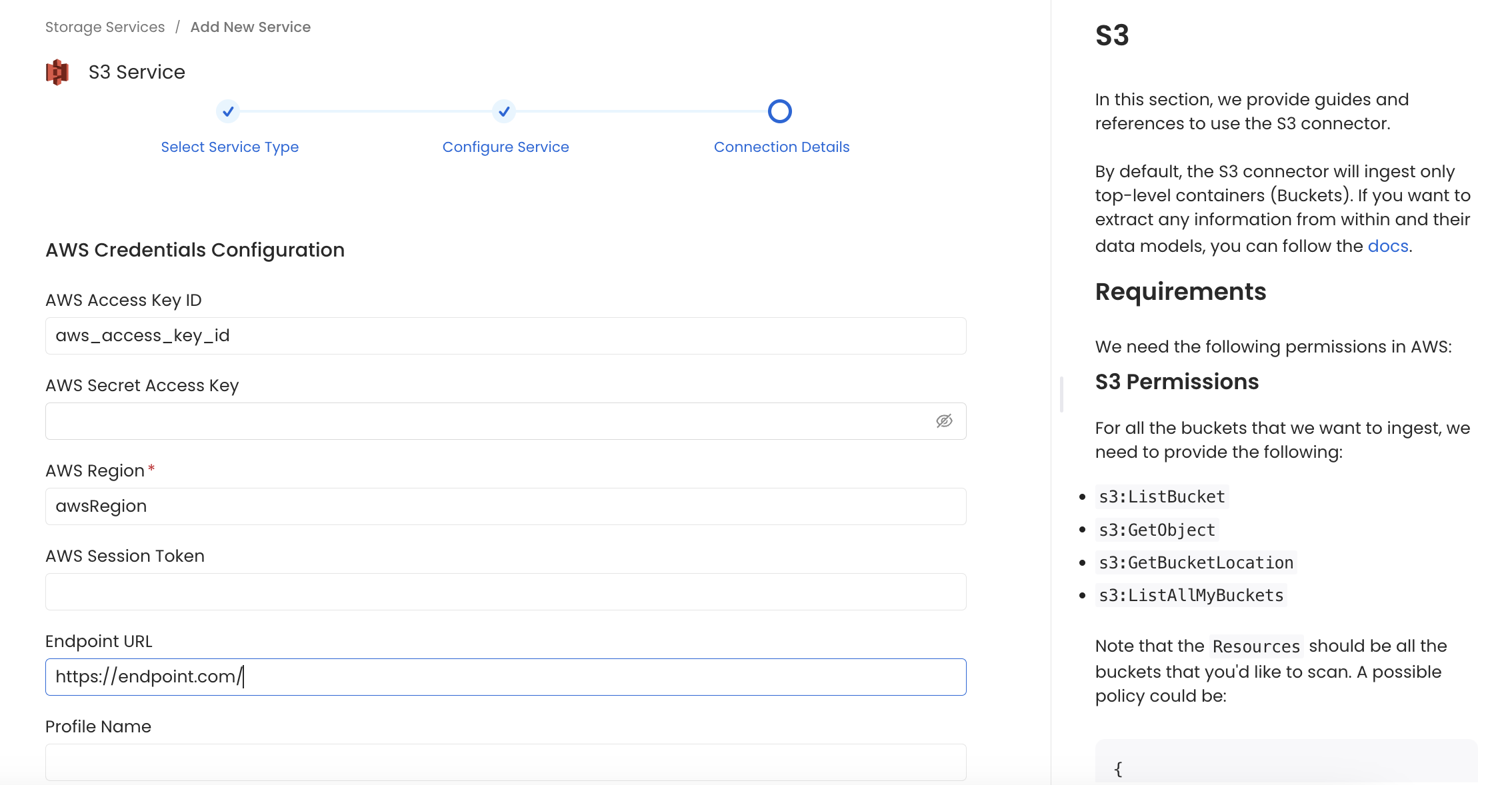
Configure the service connection by filling the form
Connection Details
- AWS Access Key ID & AWS Secret Access Key: When you interact with AWS, you specify your AWS security credentials to verify who you are and whether you have permission to access the resources that you are requesting. AWS uses the security credentials to authenticate and authorize your requests (docs).
Access keys consist of two parts: An access key ID (for example, AKIAIOSFODNN7EXAMPLE), and a secret access key (for example, wJalrXUtnFEMI/K7MDENG/bPxRfiCYEXAMPLEKEY).
You must use both the access key ID and secret access key together to authenticate your requests.
You can find further information on how to manage your access keys here.
- AWS Region: Each AWS Region is a separate geographic area in which AWS clusters data centers (docs).
As AWS can have instances in multiple regions, we need to know the region the service you want reach belongs to.
Note that the AWS Region is the only required parameter when configuring a connection. When connecting to the services programmatically, there are different ways in which we can extract and use the rest of AWS configurations.
You can find further information about configuring your credentials here.
- AWS Session Token (optional): If you are using temporary credentials to access your services, you will need to inform the AWS Access Key ID and AWS Secrets Access Key. Also, these will include an AWS Session Token.
You can find more information on Using temporary credentials with AWS resources.
- Endpoint URL (optional): To connect programmatically to an AWS service, you use an endpoint. An endpoint is the URL of the entry point for an AWS web service. The AWS SDKs and the AWS Command Line Interface (AWS CLI) automatically use the default endpoint for each service in an AWS Region. But you can specify an alternate endpoint for your API requests.
Find more information on AWS service endpoints.
- Console Endpoint URL (optional): The Console Endpoint URL is used to generate clickable links in the OpenMetadata UI that direct users to the appropriate web console for viewing buckets and objects.
For AWS S3: Leave this field empty. OpenMetadata will automatically generate the correct AWS console URLs based on your bucket's region.
For S3-compatible services (MinIO, Cloudflare R2, DigitalOcean Spaces, Wasabi, etc.): Provide the base console path where your buckets can be viewed in the web interface.
Examples:
- MinIO:
http://minio.example.com:9001/browser/ - Cloudflare R2:
https://dash.cloudflare.com/?to=/<account-id>/r2/buckets/ - DigitalOcean Spaces:
https://cloud.digitalocean.com/spaces/ - Wasabi:
https://console.wasabisys.com/#/file_manager/
OpenMetadata will append the bucket name to this URL to create direct links to your buckets.
- Profile Name: A named profile is a collection of settings and credentials that you can apply to a AWS CLI command. When you specify a profile to run a command, the settings and credentials are used to run that command. Multiple named profiles can be stored in the config and credentials files.
You can inform this field if you'd like to use a profile other than default.
Find here more information about Named profiles for the AWS CLI.
- Assume Role Arn: Typically, you use
AssumeRolewithin your account or for cross-account access. In this field you'll set theARN(Amazon Resource Name) of the policy of the other account.
A user who wants to access a role in a different account must also have permissions that are delegated from the account administrator. The administrator must attach a policy that allows the user to call AssumeRole for the ARN of the role in the other account.
This is a required field if you'd like to AssumeRole.
Find more information on AssumeRole.
When using Assume Role authentication, ensure you provide the following details:
- AWS Region: Specify the AWS region for your deployment.
- Assume Role ARN: Provide the ARN of the role in your AWS account that OpenMetadata will assume.
- Assume Role Session Name: An identifier for the assumed role session. Use the role session name to uniquely identify a session when the same role is assumed by different principals or for different reasons.
By default, we'll use the name OpenMetadataSession.
Find more information about the Role Session Name.
- Assume Role Source Identity: The source identity specified by the principal that is calling the
AssumeRoleoperation. You can use source identity information in AWS CloudTrail logs to determine who took actions with a role.
Find more information about Source Identity.
- Bucket Names (Optional): Provide the names of buckets that you would want to ingest, if you want to ingest metadata from all buckets or apply a filter to ingest buckets then leave this field empty.
6. Test the Connection
Once the credentials have been added, click on Test Connection and Save the changes.
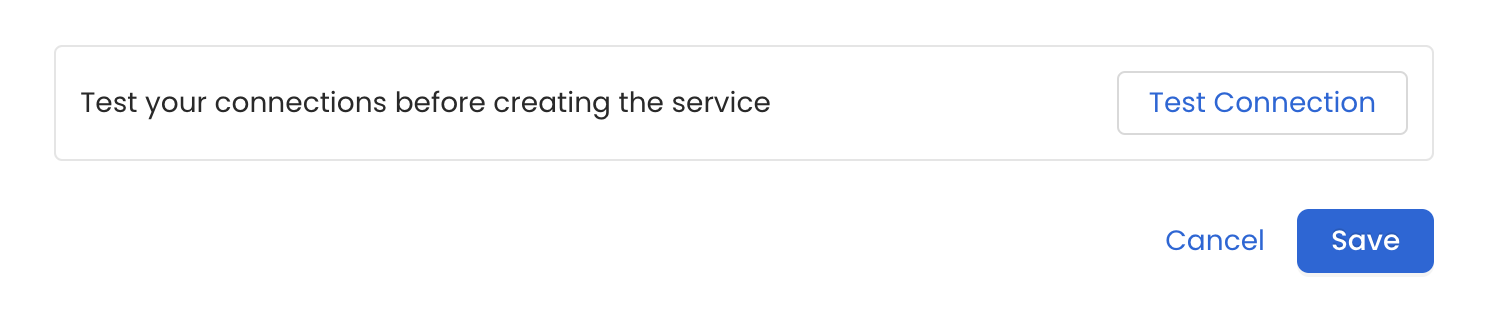
Test the connection and save the Service
7. Configure Metadata Ingestion
In this step we will configure the metadata ingestion pipeline, Please follow the instructions below
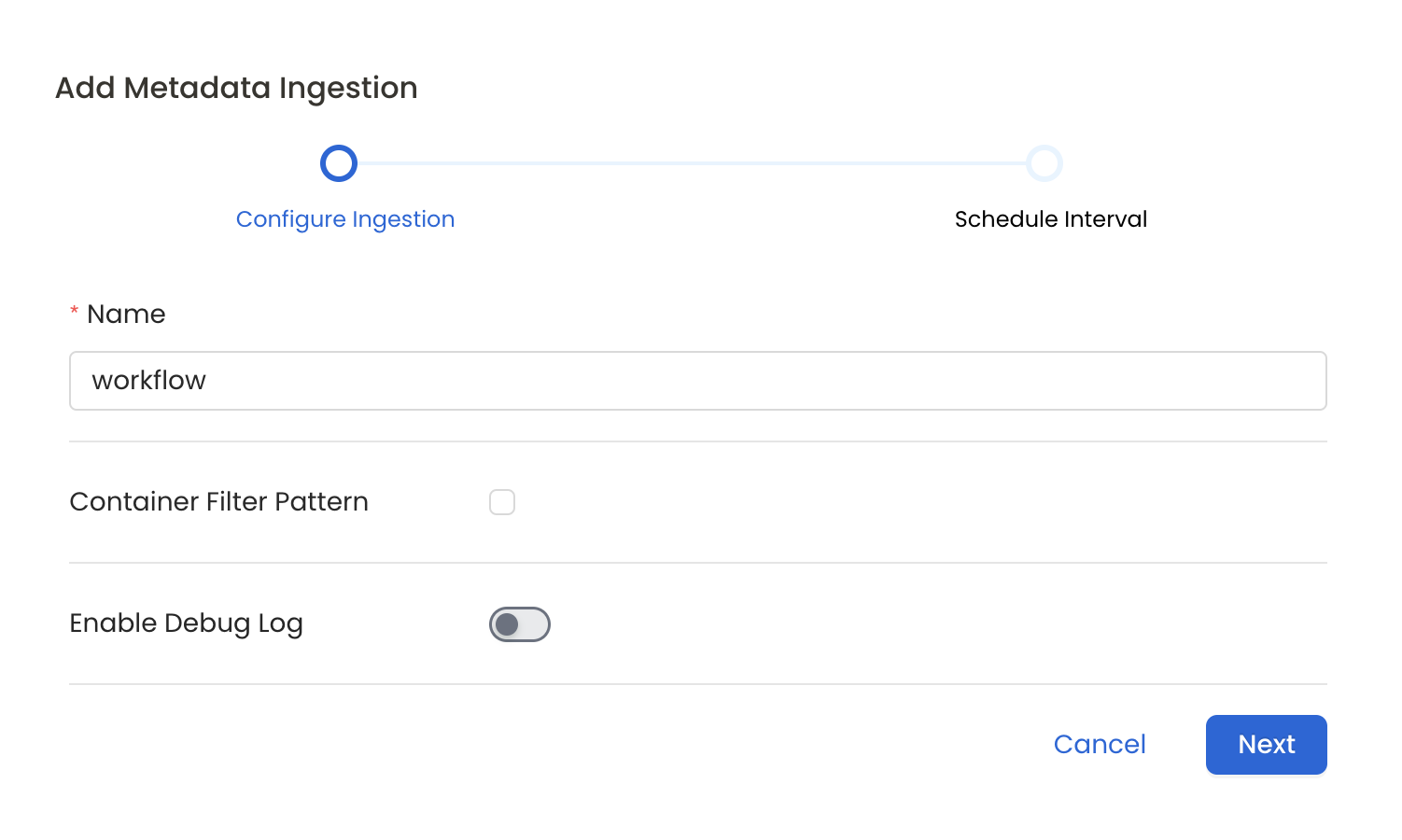
Configure Metadata Ingestion Page
Metadata Ingestion Options
- Name: This field refers to the name of ingestion pipeline, you can customize the name or use the generated name.
- Container Filter Pattern (Optional): To control whether to include a container as part of metadata ingestion.
- Include: Explicitly include containers by adding a list of comma-separated regular expressions to the Include field. OpenMetadata will include all containers with names matching one or more of the supplied regular expressions. All other containers will be excluded.
- Exclude: Explicitly exclude containers by adding a list of comma-separated regular expressions to the Exclude field. OpenMetadata will exclude all containers with names matching one or more of the supplied regular expressions. All other containers will be included.
- Enable Debug Log (toggle): Set the Enable Debug Log toggle to set the default log level to debug.
- Storage Metadata Config Source: Here you can specify the location of your global manifest
openmetadata_storage_manifest.jsonfile. It can be located in S3, a local path or HTTP.
8. Schedule the Ingestion and Deploy
Scheduling can be set up at an hourly, daily, weekly, or manual cadence. The timezone is in UTC. Select a Start Date to schedule for ingestion. It is optional to add an End Date.
Review your configuration settings. If they match what you intended, click Deploy to create the service and schedule metadata ingestion.
If something doesn't look right, click the Back button to return to the appropriate step and change the settings as needed.
After configuring the workflow, you can click on Deploy to create the pipeline.
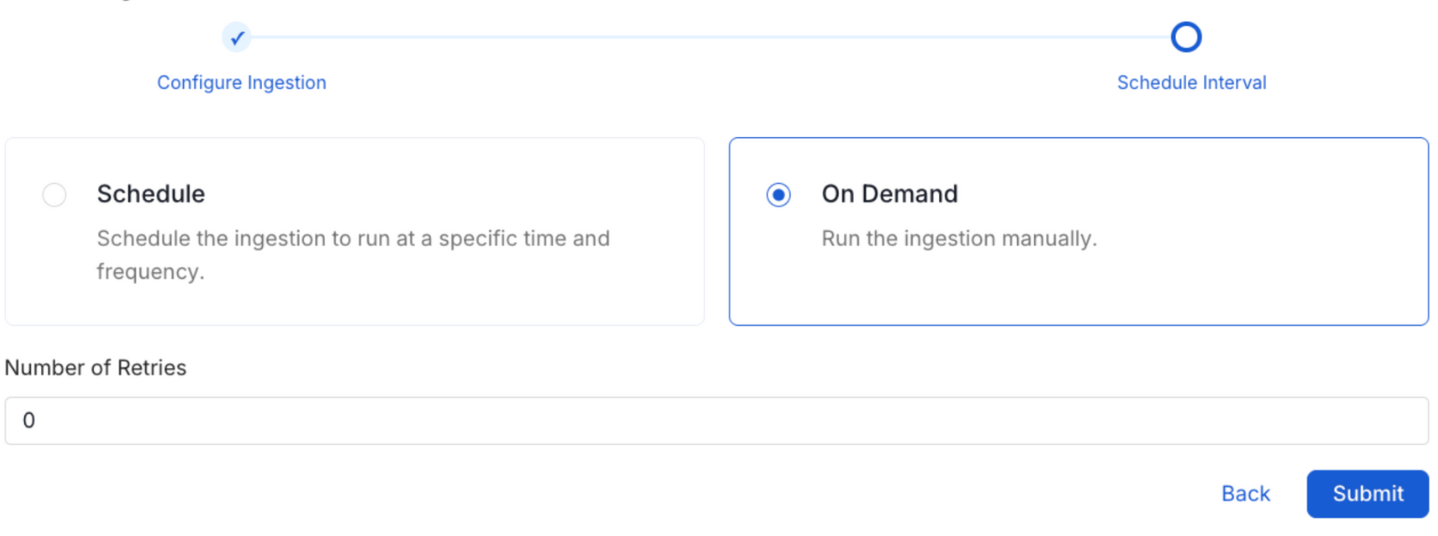
Schedule the Ingestion Pipeline and Deploy
9. View the Ingestion Pipeline
Once the workflow has been successfully deployed, you can view the Ingestion Pipeline running from the Service Page.
View the Ingestion Pipeline from the Service Page
If AutoPilot is enabled, workflows like usage tracking, data lineage, and similar tasks will be handled automatically. Users don’t need to set up or manage them - AutoPilot takes care of everything in the system.