Configure dbt workflow from OpenMetadata UI
Learn how to configure the dbt workflow from the UI to ingest dbt data from your data sources.
- OpenMetadata supports both dbt Core and dbt Cloud for databases. After metadata ingestion, OpenMetadata extracts model information from dbt and integrates it accordingly.
- Additionally, dbt Cloud supports executing models directly. OpenMetadata enables ingestion of these executions as a Pipeline Service for enhanced tracking and visibility.
UI Configuration
Once the metadata ingestion runs correctly and we are able to explore the service Entities, we can add the dbt information.
This will populate the dbt tab from the Table Entity Page.
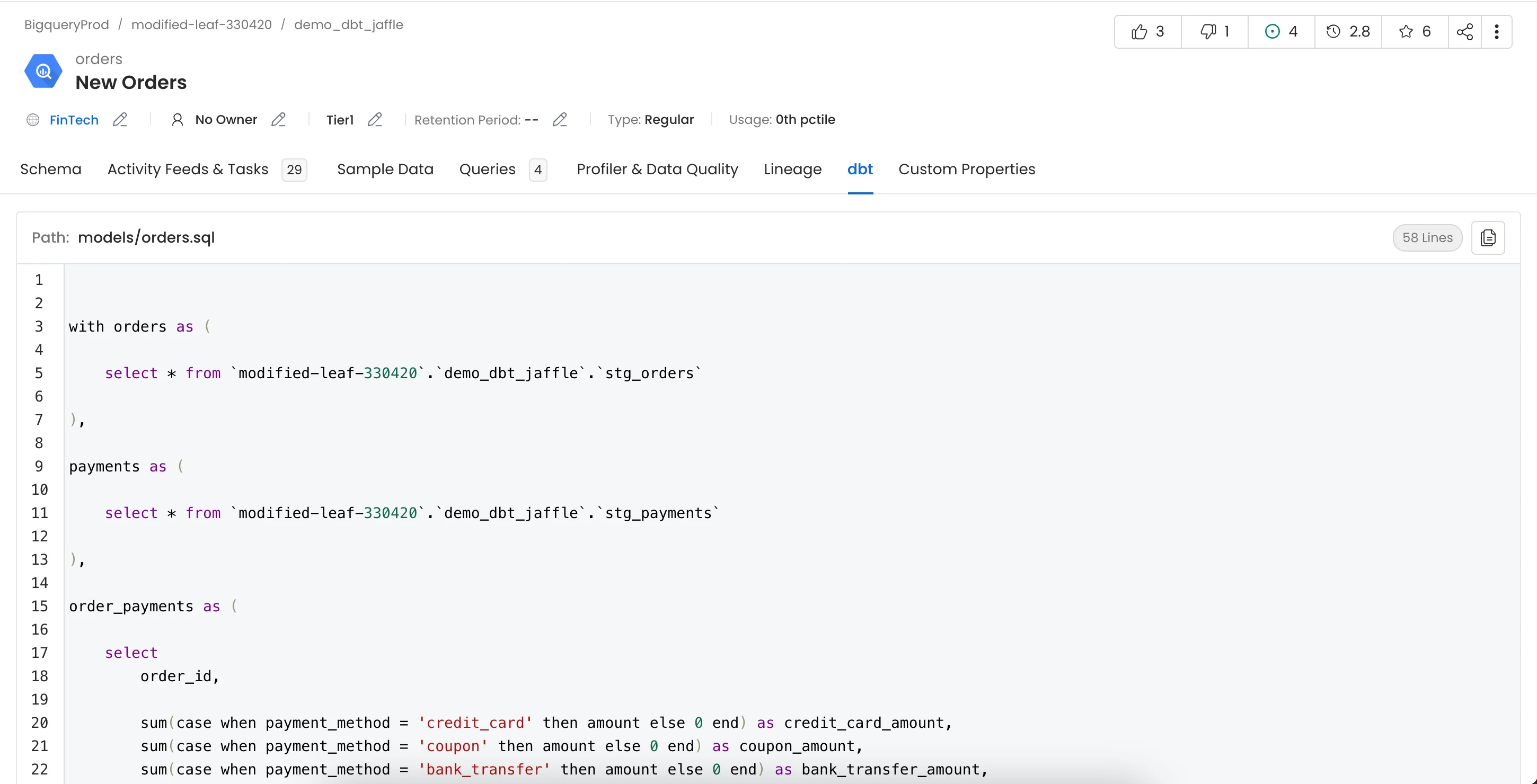
dbt
We can create a workflow that will obtain the dbt information from the dbt files and feed it to OpenMetadata. The dbt Ingestion will be in charge of obtaining this data.
1. Add a dbt Ingestion
From the Service Page, go to the Ingestions tab to add a new ingestion and click on Add dbt Ingestion.
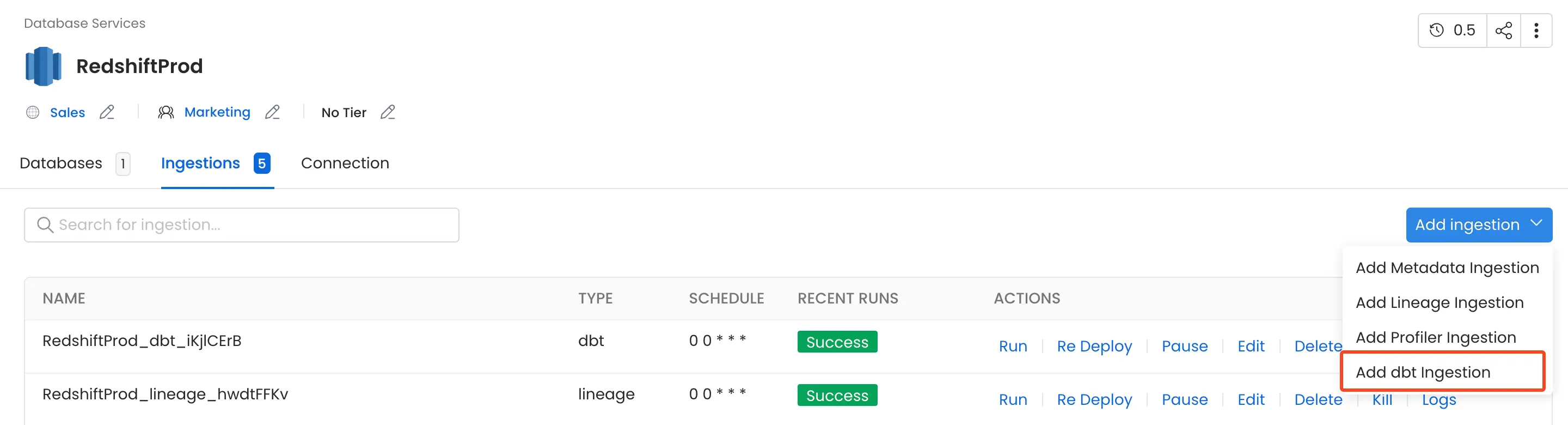
Add dbt Ingestion
2. Configure the dbt Ingestion
Here you can enter the configuration required for OpenMetadata to get the dbt files (manifest.json, catalog.json and run_results.json) required to extract the dbt metadata. Select any one of the source from below from where the dbt files can be fetched:
Only the manifest.json file is compulsory for dbt ingestion.
1. AWS S3 Buckets
OpenMetadata connects to the AWS s3 bucket via the credentials provided and scans the AWS s3 buckets for manifest.json, catalog.json and run_results.json files.
The name of the s3 bucket and prefix path to the folder in which the dbt files are stored can be provided. In the case where these parameters are not provided all the buckets are scanned for the files.
Follow the link here for instructions on setting up multiple dbt projects.
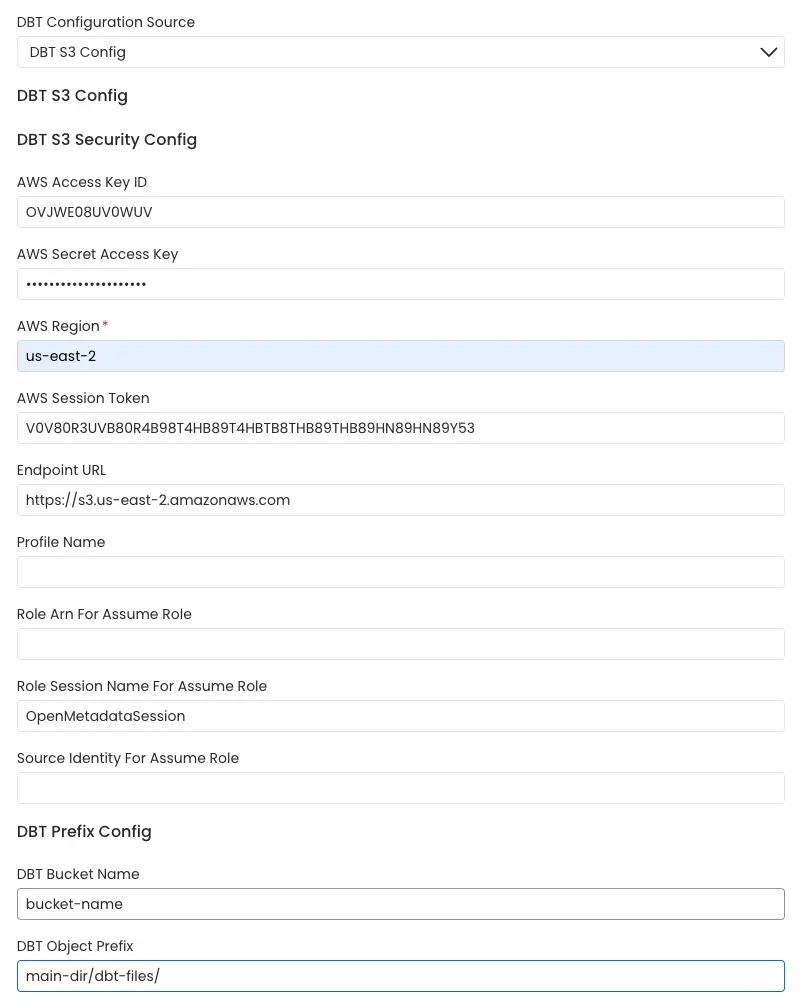
AWS S3 Bucket Config
2. Google Cloud Storage Buckets
OpenMetadata connects to the GCS bucket via the credentials provided and scans the gcp buckets for manifest.json, catalog.json and run_results.json files.
The name of the GCS bucket and prefix path to the folder in which the dbt files are stored can be provided. In the case where these parameters are not provided all the buckets are scanned for the files.
GCS credentials can be stored in two ways:
1. Entering the credentials directly into the form
Follow the link here for instructions on setting up multiple dbt projects.

GCS Bucket config
2. Entering the path of file in which the GCS bucket credentials are stored.
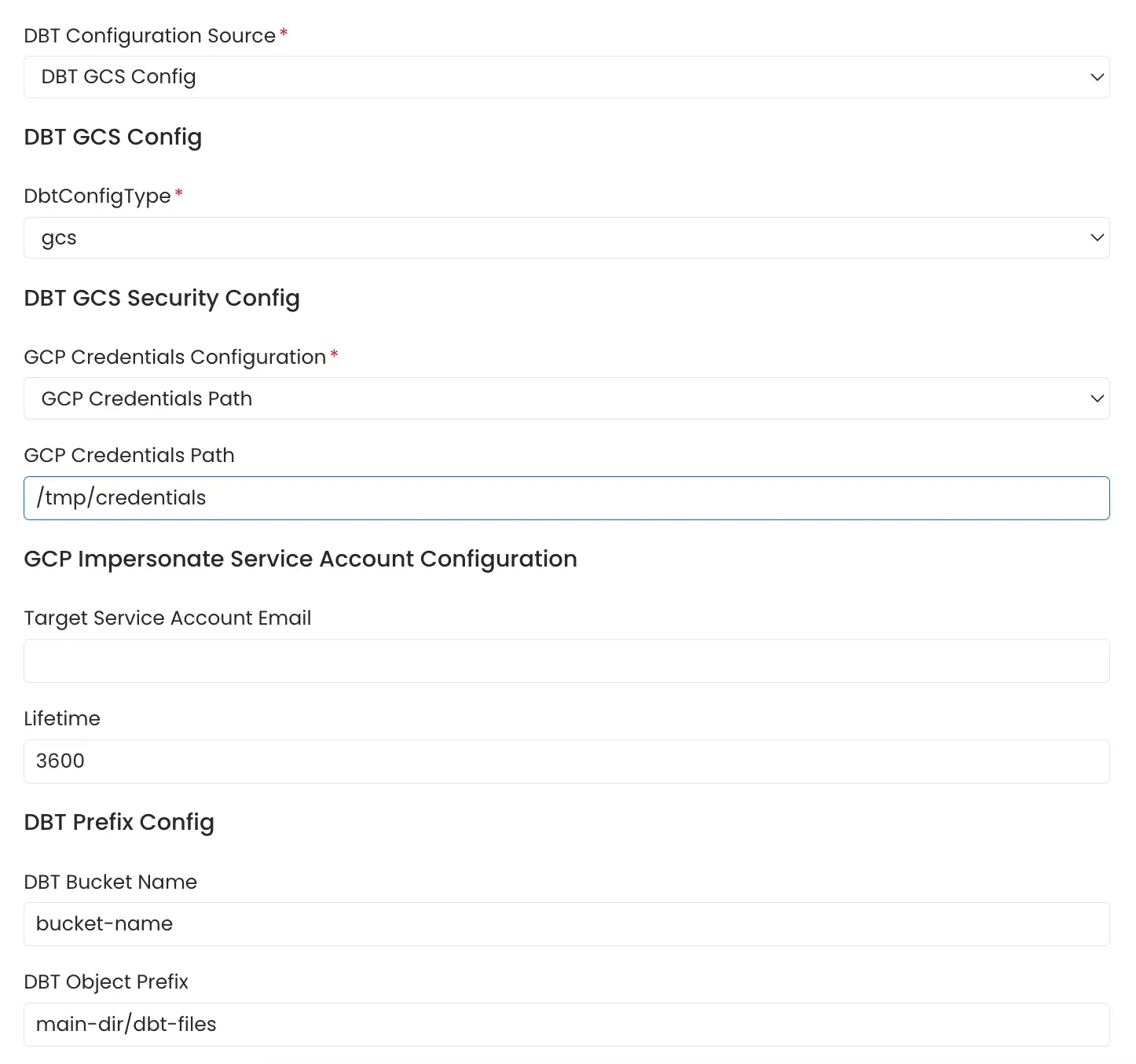
GCS Bucket Path Config
For more information on Google Cloud Storage authentication click here.
3. Azure Storage Buckets
OpenMetadata connects to the Azure Storage service via the credentials provided and scans the AWS s3 buckets for manifest.json, catalog.json and run_results.json files.
The name of the s3 bucket and prefix path to the folder in which the dbt files are stored can be provided. In the case where these parameters are not provided all the buckets are scanned for the files.
Follow the link here for instructions on setting up multiple dbt projects.
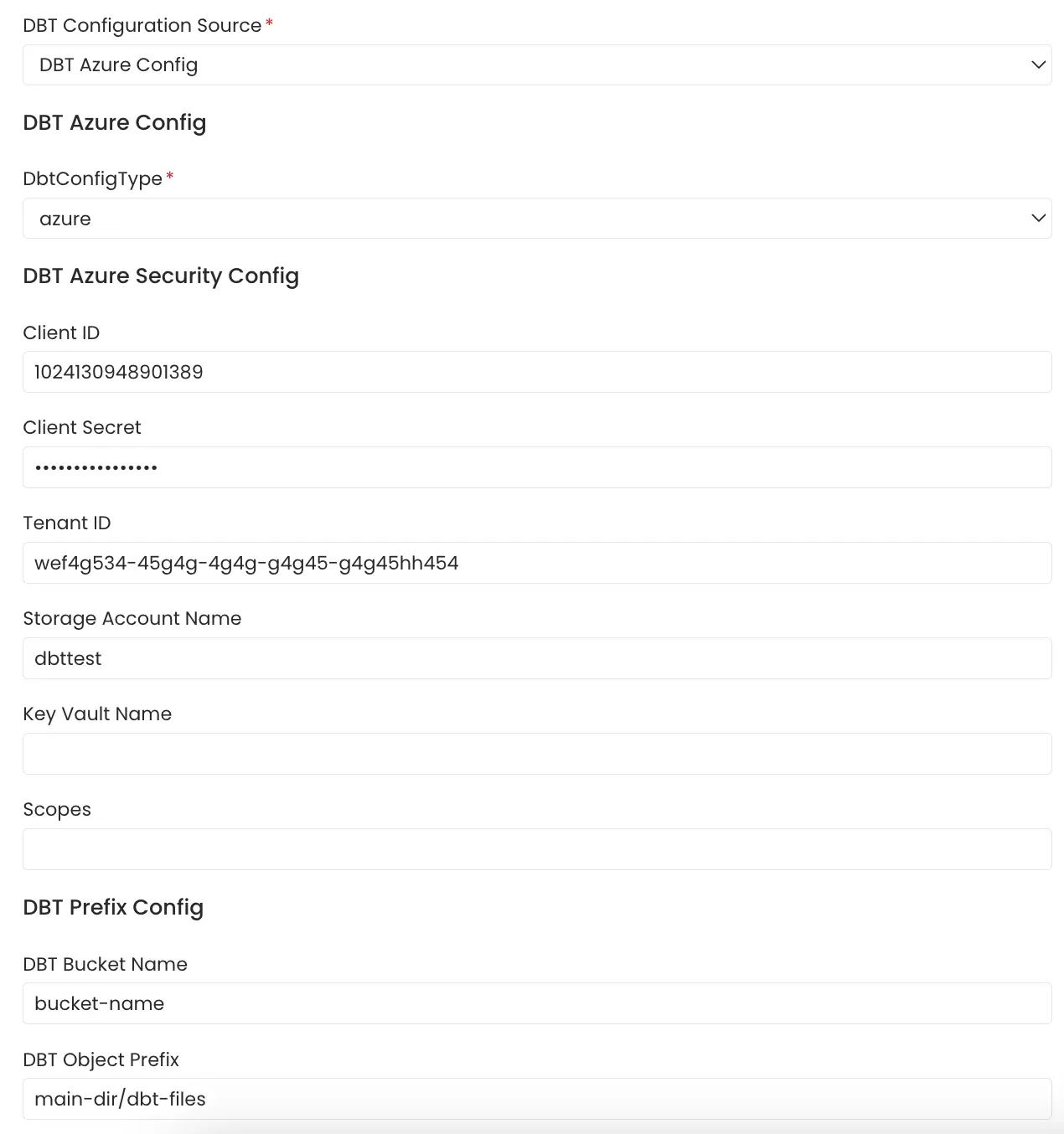
Azure Storage Config
4. Local Storage
Path of the manifest.json, catalog.json and run_results.json files stored in the local system or in the container in which openmetadata server is running can be directly provided.
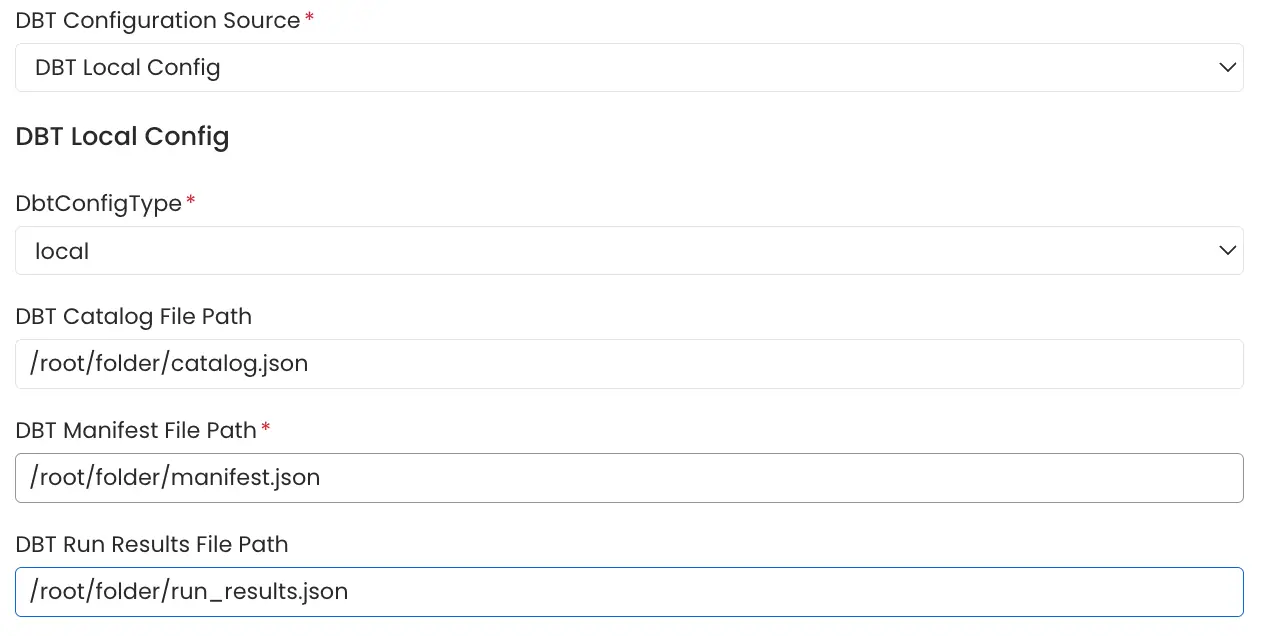
Local Storage Config
5. File Server
File server path of the manifest.json, catalog.json and run_results.json files stored on a file server directly provided.
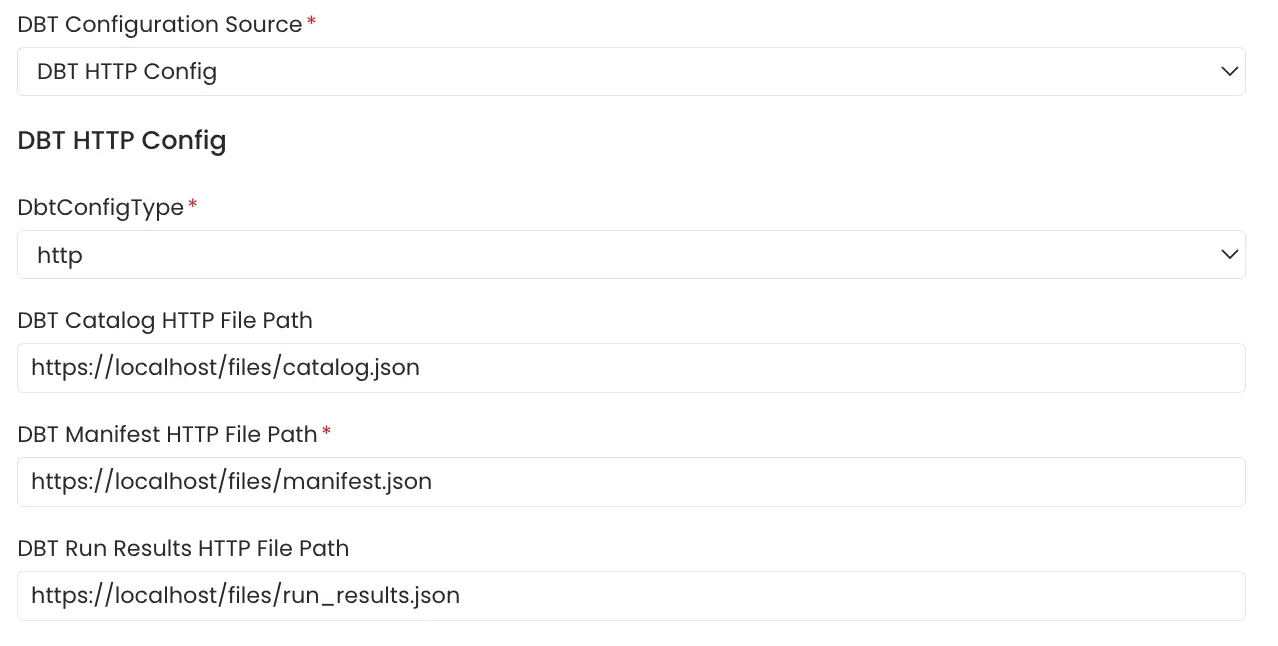
File Server Config
6. dbt Cloud
Click on the the link here for getting started with dbt cloud account setup if not done already. The APIs need to be authenticated using an Authentication Token. Follow the link here to generate an authentication token for your dbt cloud account.
The Account Viewer permission is the minimum requirement for the dbt cloud token.
The dbt Cloud workflow leverages the dbt Cloud v2 APIs to retrieve dbt run artifacts (manifest.json, catalog.json, and run_results.json) and ingest the dbt metadata.
It uses the /runs API to obtain the most recent successful dbt run, filtering by account_id, project_id and job_id if specified. The artifacts from this run are then collected using the /artifacts API.
Refer to the code here
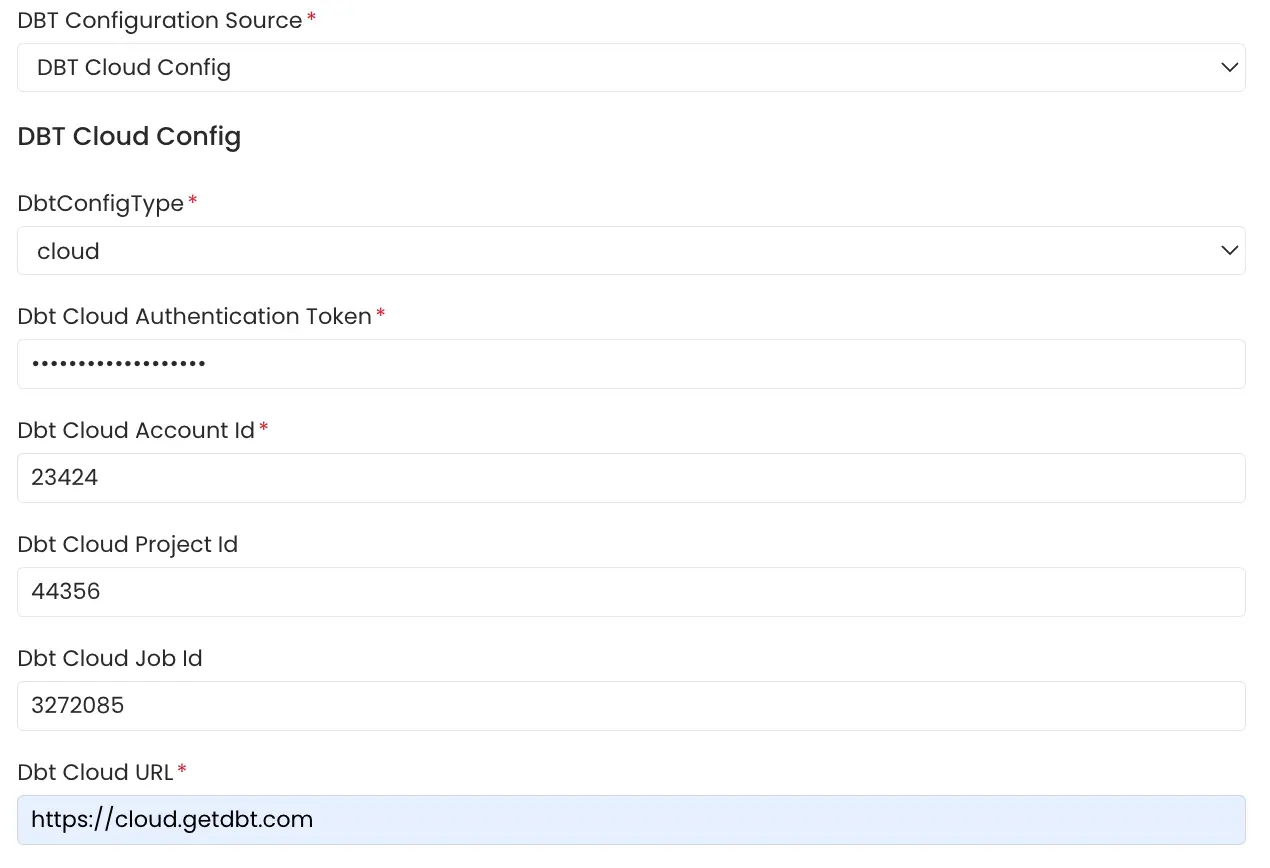
dbt Cloud config
The fields for Dbt Cloud Account Id, Dbt Cloud Project Id and Dbt Cloud Job Id should be numeric values.
To know how to get the values for Dbt Cloud Account Id, Dbt Cloud Project Id and Dbt Cloud Job Id fields check here.
3. Schedule and Deploy
After clicking Next, you will be redirected to the Scheduling form. This will be the same as the Metadata Ingestion. Select your desired schedule and click on Deploy to find the lineage pipeline being added to the Service Ingestions.
Schedule dbt ingestion pipeline